Research
Current Science
The traditional approach to microbiome research, which is primarily correlative, has its limitations in uncovering the mechanistic roles of the microbiome in health and disease. At the Zarrinpar Lab, we aim to shift from this correlative lens to a mechanistic one, recognizing the microbiome as a dynamic, ever-fluctuating system. These microbiome fluctuations, we believe, are important biological signals influencing health outcomes. We strive to understand the functional roles and timing of specific microbiome features and the difference between those causing disease versus those resulting from it. Deciphering these complexities is a crucial step towards effectively leveraging the gut microbiome for disease diagnosis, prevention, and cure.
Our Impact
At the Zarrinpar Lab, we delve into the pivotal role the gut plays in host physiology, specifically focusing on the interplay between the luminal environment – including the microbiome and its secondary metabolites – and intestinal cells. This intricate interaction establishes a physiological equilibrium that extends beyond the gut. However, any disruption to this homeostasis can trigger metabolic irregularities and an increased susceptibility to inflammation in other target organs.
Our research targets the dynamic influence of diet composition and daily eating patterns on this equilibrium, particularly how they alter the cyclical dynamics of the gut microbiome, intestinal gene expression, and gut signaling. By understanding and manipulating these systems, we aim to unveil novel pathways influencing host metabolism. Our ultimate goal is to translate our findings from rodent studies to humans. We seek to accomplish this through longitudinal observational studies and clinical trials, in the hope that our research will pave the way for impactful advancements in the understanding and treatment of health conditions linked to the gut microbiome.
Our Plan
1) To engineer effective microbiome-targeted therapeutics.
In our quest to develop more effective microbiome-targeted therapeutics, the Zarrinpar Lab has developed a method using engineered native bacteria, opening new frontiers in our understanding of the gut microbiome’s impact on host physiology. In a recent study, we demonstrated that these engineered bacteria can stably colonize the gut, bring about functional changes in the body, alter specific metabolites in the bloodstream, and even ameliorate certain disease conditions in conventionally raised wildtype mice.
With this new technique, we have started to uncover the profound influence of microbiome functions and the fluctuating/circadian luminal environment on host inflammation, metabolism, cancer development, and other diseases. Crucially, these advances enable us to design novel therapeutic strategies with the potential to cure disease, while taking into account the wide variability caused by diet and circadian rhythms.
2) To understand the contribution of the bacterial bile acid biotransformations on physiological processes and disease state
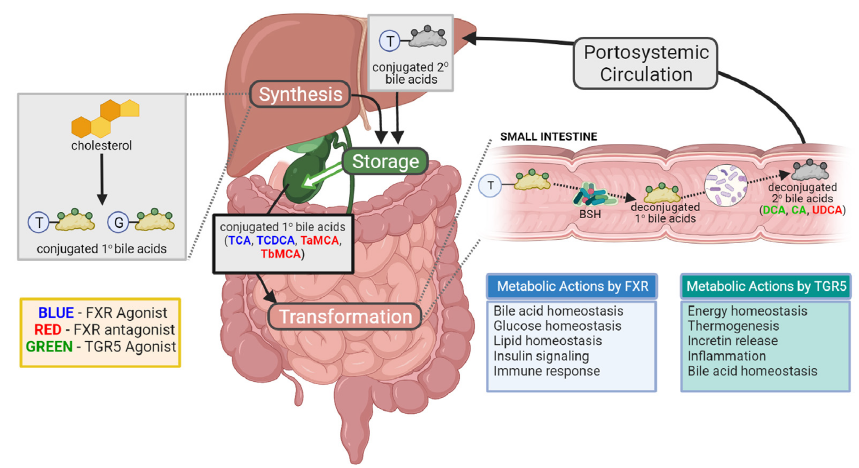
Pioneering the exploration of bacterial bile acid transformations on various physiological processes and disease states, the Zarrinpar Lab has developed a unique engineered bacterium. This bacterium, capable of manipulating luminal bile acids, serves as a powerful tool to investigate how these transformations affect an array of diseases, such as type 2 diabetes, atherosclerosis, colorectal cancer, aging, polycystic ovary syndrome, and inflammatory bowel disease. By focusing on these bacterial transformations, our projects will provide a more in-depth, mechanistic understanding of the gut microbiome’s communication with the host. This, in turn, will further our knowledge of how the host adapts its physiological processes in response to these microbial signals, opening up new frontiers for targeted therapeutic strategies.
3) To understand the microbe-host relationship and coevolution as a dynamic state
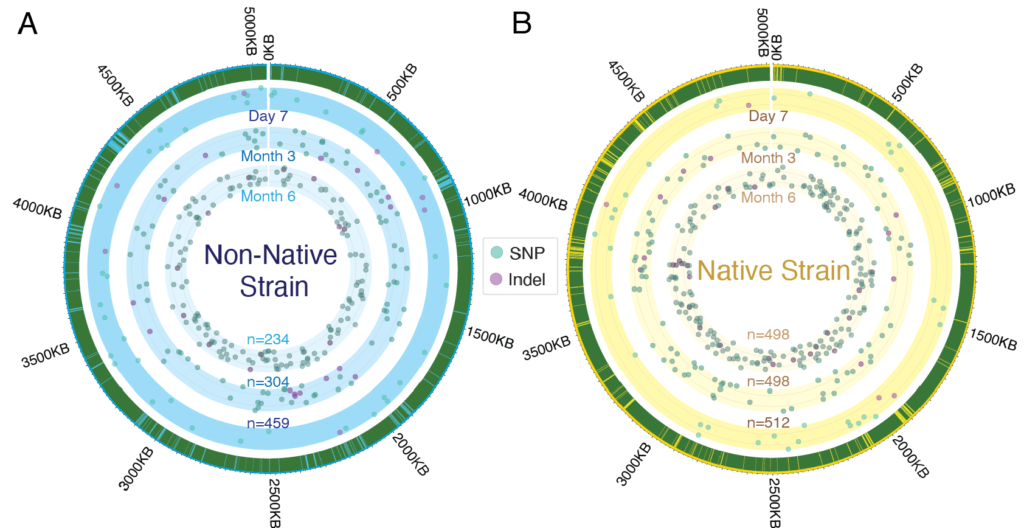
Deciphering the dynamic nature of the gut microbiome is a vital component of our research at Zarrinpar Lab. Our studies have uncovered how temporal oscillations in microbiome composition, underappreciated until now, significantly impact study results and have vital implications for host health. Our work delves into the dynamic aspects of the gut microbiome: from daily fluctuations in bacterial composition, functional potential, and the availability of host receptors to alterations in bacterial genomes over time. These temporal aspects of the microbiome may explain the challenge in defining a ‘normal’ gut microbiome due to inherent variability. We have also focused on understanding how bacteria adapt within the host microbiome, a factor of immense importance for the success of live bacterial therapeutics. Furthermore, we’ve shown that the time of sample collection in microbiome studies can significantly affect the conclusions drawn. Like trying to measure sea level without considering tides, disregarding these microbiome dynamics can obscure the true impact of interventions, highlighting the need for standardization across studies.
4) To use what we have learned about the gut microbiome, gut physiology, and circadian rhythms to improve patient care
At Zarrinpar Lab, we’re using our knowledge of the gut microbiome, gut physiology, and circadian rhythms to revolutionize patient care. Our research ambitiously pursues personalized treatment approaches for obesity, metabolic disorders, and other chronic gastrointestinal disorders. We’ve delved into using nutrigenetic profiles to guide dietary therapy and investigating the role of microRNAs as potential fatty liver disease biomarkers. We’ve also probed the effects of obesity-inducing medications on weight loss outcomes. We’re spearheading the use of microbial DNA from low biomass samples as potential diagnostic or prognostic tools. Furthermore, we’re determining how circadian rhythms can be integrated into diagnostic and therapeutic medicine. Through this, we aim to enhance healthcare outcomes by bringing advanced research into clinical application, ultimately benefiting patients.


